 We will use the same notation, and additionally use \(f\) to refer to the layer after the BN
layer.
We will use the same notation, and additionally use \(f\) to refer to the layer after the BN
layer.June 24th, 2025
In this note I want to share a method of computing an analytical gradient for Batch Normalization that I found to be most clear and understandable, to me.
I was prompted to do this while working on assignment 2 from CS231n, where you are asked to derive and implement the backward pass for batch normalization from scratch. To my understanding there are two ways to do this:
I tried to take the analytical derivative approach because I found that it made the process most intuitive, given that you understand the chain rule. There are a few other blog posts on this topic, one uses the computational graph approach, two and three use the analytical gradient derivative. This note will aim to be more explanatory in both the maths and the code. We will assume you understand BatchNorm, Calculus 1 derivative rules, and the conceptual different between a derivative and a partial derivative.
Some advice in reading this: it takes some time to load the priors of Batch Norm into your head, so skipping parts or scrolling through the note will probably confuse you and serve as discouragement. You will get the most out of this by following along from the beginning and writing down your understanding as you go, stopping if you get confused to rectify that confusion, and repeating.
Finally, I'm aware that BatchNorm isn't really used in practice very much anymore, in favour of alternatives such as LayerNorm and GroupNorm, but I believe deriving and implementing the backward pass for BatchNorm is a valuable exercise for newcomers to deep learning because the input effects the output in several different ways, making for a complicated practice problem.
Please write to me with any feedback, errata, or thoughts via email or X.
Before diving into the batch normalization gradient derivation, let's review the multivariable chain rule since it's the foundation of our approach.
Consider a function \(u(x,y)\) where both \(x\) and \(y\) are themselves functions of other variables, say \(x(r,t)\) and \(y(r,t)\). When we want to find how \(u\) changes with respect to \(r\), we need to account for all the ways that \(r\) influences \(u\). Since \(r\) affects \(u\) through both \(x\) and \(y\), the chain rule tells us: \[\frac{\partial u}{\partial r} = \frac{\partial u}{\partial x} \cdot \frac{\partial x}{\partial r} + \frac{\partial u}{\partial y} \cdot \frac{\partial y}{\partial r}\] This principle extends naturally to functions with more variables. The key insight is that we sum up all the "pathways" through which our variable of interest can influence the final output. In batch normalization, this becomes particularly important because the input affects the output through multiple computational paths.
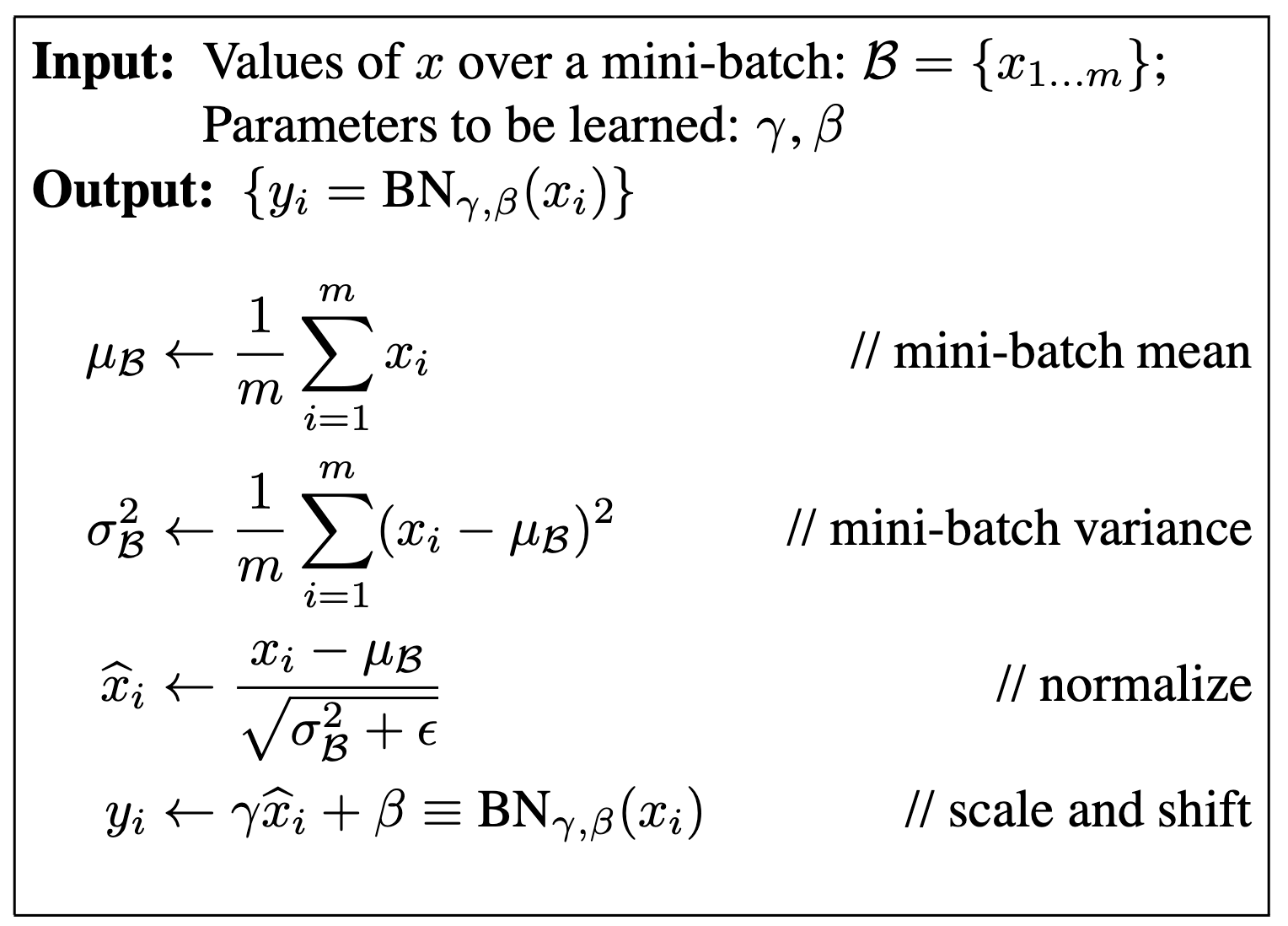
From the original paper BatchNorm is given as:
 We will use the same notation, and additionally use \(f\) to refer to the layer after the BN
layer.
We will use the same notation, and additionally use \(f\) to refer to the layer after the BN
layer.
We start by deriving the gradient of \(f\) with respect to the a single input \(x_i\). Using the chain rule, we have:
\[\frac{df}{dx_i} = \sum_j \left( \frac{\partial f}{\partial y_j} \cdot \frac{\partial y_j}{\partial x_i} \right)\] where:
You may be confused by the summation, but the reason for it is that a single input \(x_i\) affects every single output \(y_j\) in the batch, not just \(y_i\). This happens because batch normalization uses shared statistics (mean and variance) computed across the entire batch. More specifically, \(x_i\) influences each \(y_j\) through three distinct pathways:
Since we want to find how \(f\) changes with respect to \(x_i\), we need to sum up \(x_i\)'s influence on all the outputs \(y_j\) that contribute to \(f\). This is exactly what the summation \(\sum_j\) captures.
Now let's break down the effect of a single input \(x_i\) on a single output \(y_j\) \((\frac{\partial y_j}{\partial x_i})\) using the very same method from the chain rule refresher above: \[\frac{\partial y_j}{\partial x_i} = \underbrace{\textcolor{green}{\frac{\partial y_j}{\partial \hat{x}_j} \cdot \frac{\partial \hat{x}_j}{\partial x_i}}}_{\text{direct path}} + \underbrace{\textcolor{blue}{\frac{\partial y_j}{\partial \hat{x}_j} \cdot \frac{\partial \hat{x}_j}{\partial \mu_B}} \cdot \textcolor{blue}{\frac{\partial \mu_B}{\partial x_i}}}_{\text{mean path}} + \underbrace{\textcolor{red}{\frac{\partial y_j}{\partial \hat{x}_j} \cdot \frac{\partial \hat{x}_j}{\partial \sigma_B^2}} \cdot \textcolor{red}{\frac{\mathrm{d} \sigma_B^2}{\mathrm{d} x_i}}}_{\text{variance path}}\] Factoring out \(\frac{\partial y_j}{\partial \hat{x}_j}\) we get: \[\frac{\partial y_j}{\partial x_i} = \frac{\partial y_j}{\partial \hat{x}_j} \left( \underbrace{\textcolor{green}{\frac{\partial \hat{x}_j}{\partial x_i}}}_{\text{direct}} + \underbrace{\textcolor{blue}{\frac{\partial \hat{x}_j}{\partial \mu_B} \cdot \frac{\partial \mu_B}{\partial x_i}}}_{\text{mean}} + \underbrace{\textcolor{red}{\frac{\partial \hat{x}_j}{\partial \sigma_B^2} \cdot \frac{\mathrm{d} \sigma_B^2}{\mathrm{d} x_i}}}_{\text{variance}} \right)\]
Now let's compute each pathway step by step.
For the direct path, we need to compute \(\textcolor{green}{\frac{\partial \hat{x}_j}{\partial x_i}}\). Recall that \(\hat{x}_j = \frac{x_j - \mu_B}{\sqrt{\sigma_B^2 + \epsilon}}\). When we take the partial derivative with respect to \(x_i\), we only get a non-zero result when \(j = i\) (since \(x_j\) doesn't directly depend on \(x_i\) unless they're the same variable).
\[\textcolor{green}{\frac{\partial \hat{x}_j}{\partial x_i}} = \begin{cases} \frac{1}{\sqrt{\sigma_B^2 + \epsilon}} & \text{if } i = j \\ 0 & \text{if } i \neq j \end{cases}\]
This can be written compactly using the Kronecker delta \(\delta_{ij}\), which equals 1 when \(i = j\) and 0 otherwise:
\[\textcolor{green}{\frac{\partial \hat{x}_j}{\partial x_i}} = \frac{\delta_{ij}}{\sqrt{\sigma_B^2 + \epsilon}}\]
For the mean path, we need two derivatives: \(\textcolor{blue}{\frac{\partial \hat{x}_j}{\partial \mu_B}}\) and \(\textcolor{blue}{\frac{\partial \mu_B}{\partial x_i}}\). First, \(\textcolor{blue}{\frac{\partial \hat{x}_j}{\partial \mu_B}}\): \[\hat{x}_j = \frac{x_j - \mu_B}{\sqrt{\sigma_B^2 + \epsilon}}\] When taking the partial derivative with respect to \(\mu_B\), both \(x_j\) and \(\sqrt{\sigma_B^2 + \epsilon}\) are treated as constants. We're essentially differentiating \((x_j - \mu_B)\) with respect to \(\mu_B\), which gives us \(-1\), then multiplying by the constant coefficient \(\frac{1}{\sqrt{\sigma_B^2 + \epsilon}}\):
\[\textcolor{blue}{\frac{\partial \hat{x}_j}{\partial \mu_B}} = \frac{-1}{\sqrt{\sigma_B^2 + \epsilon}}\] Next, \(\textcolor{blue}{\frac{\partial \mu_B}{\partial x_i}}\): \[\mu_B = \frac{1}{N} \sum_{k=1}^N x_k = \frac{1}{N}(x_1 + x_2 + \cdots + x_i + \cdots + x_N)\]
When we expand the summation and take the partial derivative with respect to \(x_i\), all terms \(x_k\) where \(k \neq i\) are constants and disappear. Only the \(x_i\) term remains, which has derivative \(1\), leaving us with the coefficient: \[\textcolor{blue}{\frac{\partial \mu_B}{\partial x_i}} = \frac{1}{N}\]
For the variance path, we need two derivatives: \(\textcolor{red}{\frac{\partial \hat{x}_j}{\partial \sigma_B^2}}\) and \(\textcolor{red}{\frac{\partial \sigma_B^2}{\partial x_i}}\).
First, \(\textcolor{red}{\frac{\partial \hat{x}_j}{\partial \sigma_B^2}}\): \[\hat{x}_j = (x_j - \mu_B)(\sigma_B^2 + \epsilon)^{-1/2}\]
When taking the partial derivative with respect to \(\sigma_B^2\), the term \((x_j - \mu_B)\) is treated as a constant coefficient. We're differentiating \((\sigma_B^2 + \epsilon)^{-1/2}\) with respect to \(\sigma_B^2\) using the power rule: the exponent \(-1/2\) becomes the coefficient, and we subtract 1 from the exponent to get \(-3/2\):
\[\textcolor{red}{\frac{\partial \hat{x}_j}{\partial \sigma_B^2}} = (x_j - \mu_B) \cdot \left(-\frac{1}{2}\right)(\sigma_B^2 + \epsilon)^{-3/2} = -\frac{(x_j - \mu_B)}{2(\sigma_B^2 + \epsilon)^{3/2}}\] Next, \(\textcolor{red}{\frac{d \sigma_B^2}{d x_i}}\):
This requires a total derivative (not a partial derivative) because \(\sigma_B^2\) depends on \(x_i\) in two ways: directly through the \(x_i\) term when \(k=i\) in the summation, and indirectly through \(\mu_B\) (since \(\mu_B\) itself depends on all \(x_k\) including \(x_i\)). The other derivatives we computed earlier were partial derivatives because they only had one pathway of dependence. \[\sigma_B^2 = \frac{1}{N} \sum_{k=1}^N (x_k - \mu_B)^2\] Using the chain rule for total derivatives: \[\textcolor{red}{\frac{d \sigma_B^2}{d x_i}} = \frac{\partial \sigma_B^2}{\partial x_i} + \frac{\partial \sigma_B^2}{\partial \mu_B} \cdot \frac{\partial \mu_B}{\partial x_i}\]
For the first term \(\frac{\partial \sigma_B^2}{\partial x_i}\): when we treat \(\mu_B\) as constant and differentiate \((x_i - \mu_B)^2\) with respect to \(x_i\), we get \(2(x_i - \mu_B)\), multiplied by the coefficient \(\frac{1}{N}\): \[\frac{\partial \sigma_B^2}{\partial x_i} = \frac{2(x_i - \mu_B)}{N}\]
For the second term \(\frac{\partial \sigma_B^2}{\partial \mu_B}\): when we treat all \(x_k\) as constants and differentiate each \((x_k - \mu_B)^2\) with respect to \(\mu_B\), we get \(-2(x_k - \mu_B)\) for each term: \[\frac{\partial \sigma_B^2}{\partial \mu_B} = \frac{1}{N} \sum_{k=1}^N -2(x_k - \mu_B) = \frac{-2}{N} \sum_{k=1}^N (x_k - \mu_B)\]
Expanding the summation: \[\frac{\partial \sigma_B^2}{\partial \mu_B} = \frac{-2}{N} \left( \sum_{k=1}^N x_k - \sum_{k=1}^N \mu_B \right) = \frac{-2}{N} \left( \sum_{k=1}^N x_k - N\mu_B \right)\]
Since \(\mu_B = \frac{1}{N} \sum_{k=1}^N x_k\), we have \(\sum_{k=1}^N x_k = N\mu_B\), so: \[\frac{\partial \sigma_B^2}{\partial \mu_B} = \frac{-2}{N}(N\mu_B - N\mu_B) = \frac{-2}{N} \cdot 0 = 0\]
Therefore, the second term vanishes entirely, and we get: \[\textcolor{red}{\frac{d \sigma_B^2}{d x_i}} = \frac{2(x_i - \mu_B)}{N}\]
Now let's combine all the paths. Recall our original expression: \[\frac{\partial y_j}{\partial x_i} = \frac{\partial y_j}{\partial \hat{x}_j} \left( \underbrace{\textcolor{green}{\frac{\partial \hat{x}_j}{\partial x_i}}}_{\text{direct}} + \underbrace{\textcolor{blue}{\frac{\partial \hat{x}_j}{\partial \mu_B} \cdot \frac{\partial \mu_B}{\partial x_i}}}_{\text{mean}} + \underbrace{\textcolor{red}{\frac{\partial \hat{x}_j}{\partial \sigma_B^2} \cdot \frac{\mathrm{d} \sigma_B^2}{\mathrm{d} x_i}}}_{\text{variance}} \right)\]
From our calculations above, we found:
We also need \(\frac{\partial y_j}{\partial \hat{x}_j}\). Since \(y_j = \gamma \hat{x}_j + \beta\): \[\frac{\partial y_j}{\partial \hat{x}_j} = \gamma\] Substituting everything into our original expression: \[\frac{\partial y_j}{\partial x_i} = \gamma \left( \textcolor{green}{\frac{\delta_{ij}}{\sqrt{\sigma_B^2 + \epsilon}}} - \textcolor{blue}{\frac{1}{N\sqrt{\sigma_B^2 + \epsilon}}} - \textcolor{red}{\frac{(x_j - \mu_B)(x_i - \mu_B)}{N(\sigma_B^2 + \epsilon)^{3/2}}} \right)\]
We could factor out the inverse variance and make this cleaner, and thereby also make our code later cleaner, but I prefer to leave it this way because it's most intuitive to understand. This expression captures how each input \(x_i\) influences each output \(y_j\) through all three pathways we identified.
Now we can substitute our result back into the original expression to get the complete gradient of \(f\) with respect to \(x_i\): \[\frac{df}{dx_i} = \sum_j \left( \frac{\partial f}{\partial y_j} \cdot \frac{\partial y_j}{\partial x_i} \right)\] Substituting our expression for \(\frac{\partial y_j}{\partial x_i}\):
\[\frac{df}{dx_i} = \sum_j \frac{\partial f}{\partial y_j} \cdot \gamma \left( \textcolor{green}{\frac{\delta_{ij}}{\sqrt{\sigma_B^2 + \epsilon}}} - \textcolor{blue}{\frac{1}{N\sqrt{\sigma_B^2 + \epsilon}}} - \textcolor{red}{\frac{(x_j - \mu_B)(x_i - \mu_B)}{N(\sigma_B^2 + \epsilon)^{3/2}}} \right)\]
Factoring out \(\gamma\) and distributing the summation:
\[\frac{df}{dx_i} = \gamma \left[ \textcolor{green}{\sum_j \frac{\partial f}{\partial y_j} \frac{\delta_{ij}}{\sqrt{\sigma_B^2 + \epsilon}}} - \textcolor{blue}{\sum_j \frac{\partial f}{\partial y_j} \frac{1}{N\sqrt{\sigma_B^2 + \epsilon}}} - \textcolor{red}{\sum_j \frac{\partial f}{\partial y_j} \frac{(x_j - \mu_B)(x_i - \mu_B)}{N(\sigma_B^2 + \epsilon)^{3/2}}} \right]\]
Now we can simplify each colored term:
\[\frac{df}{dx_i} = \gamma \left[ \textcolor{green}{\frac{\partial f}{\partial y_i} \frac{1}{\sqrt{\sigma_B^2 + \epsilon}}} - \textcolor{blue}{\frac{1}{N\sqrt{\sigma_B^2 + \epsilon}} \sum_j \frac{\partial f}{\partial y_j}} - \textcolor{red}{\frac{(x_i - \mu_B)}{N(\sigma_B^2 + \epsilon)^{3/2}} \sum_j \frac{\partial f}{\partial y_j} (x_j - \mu_B)} \right]\]
This is our final expression for the gradient with respect to the input \(x_i\). Notice how the three pathways are still clearly visible:
That concludes the derivations for the gradient with respect to the input \(x_i\). If you got lost along the way somewhere, email me, and I'll try to help. I'd also like to hear from you if you found the explanation intuitive, I found other explanations difficult to grasp owing to my shortcomings in maths, but this worked well for me!
Next we need to find the effect of the scale parameter \(\gamma\) on \(f\), i.e. find \(\frac{df}{d\gamma}\). Again we use the chain rule. Since \(\gamma\) affects all outputs \(y_i\) in the batch, we need to sum the effect \(\gamma\) had on all \(y_i\): \[\frac{df}{d\gamma} = \sum_i \frac{\partial f}{\partial y_i} \cdot \frac{\partial y_i}{\partial \gamma}\]
We need \(\frac{\partial y_i}{\partial \gamma}\). Recall that \(y_i = \gamma \hat{x}_i + \beta\): When taking the partial derivative with respect to \(\gamma\), the term \(\beta\) disappears (it's a constant), and \(\hat{x}_i\) becomes the coefficient: \[\frac{\partial y_i}{\partial \gamma} = \hat{x}_i\] Therefore: \[\frac{df}{d\gamma} = \sum_i \frac{\partial f}{\partial y_i} \cdot \hat{x}_i\]
Similarly, for \(\frac{df}{d\beta}\): \[\frac{df}{d\beta} = \sum_i \frac{\partial f}{\partial y_i} \cdot \frac{\partial y_i}{\partial \beta}\]
We need \(\frac{\partial y_i}{\partial \beta}\). From \(y_i = \gamma \hat{x}_i + \beta\): When taking the partial derivative with respect to \(\beta\), the term \(\gamma \hat{x}_i\) disappears (it's treated as a constant), leaving us with: \[\frac{\partial y_i}{\partial \beta} = 1\] Therefore: \[\frac{df}{d\beta} = \sum_i \frac{\partial f}{\partial y_i} \cdot 1 = \sum_i \frac{\partial f}{\partial y_i}\]
These gradients are much more straightforward than the input gradient because \(\gamma\) and \(\beta\) directly scale and shift the normalized values without the complex interdependencies we saw with the shared batch statistics.
Now that we've derived the mathematical expressions for all gradients, let's see how to implement them efficiently using vectorized NumPy operations. The key challenge is understanding how the mathematical summations and individual terms map to vectorized array operations.
Here's our final vectorized implementation for reference:
def batchnorm_backward(dout, cache):
# dout: (N, D) - upstream gradients
# returns: dx (N, D), dgamma (D,), dbeta (D,)
(mean, inv_var, x, x_hat, gamma) = cache
N, D = x_hat.shape
# Gradients for learnable parameters
dgamma = np.sum(dout * x_hat, axis=0)
dbeta = np.sum(dout, axis=0)
# Input gradient: three pathway terms
# Direct contribution from x_i
term1 = dout * gamma * inv_var
# Contribution from mean (mu)
term2 = -gamma * (1. / N) * inv_var * np.sum(dout, axis=0)
# Contribution from variance (sigma^2)
term3 = -gamma * (1. / N) * (x-mean) * inv_var**3 * np.sum(dout * (x - mean), axis=0)
dx = term1 + term2 + term3
return dx, dgamma, dbetaWe'll break down how each mathematical expression maps to vectorized code.
\[\frac{df}{dx_i} = \textcolor{green}{\frac{\partial f}{\partial y_i} \frac{\gamma}{\sqrt{\sigma_B^2 + \epsilon}}} - \frac{\gamma}{N\sqrt{\sigma_B^2 + \epsilon}} \sum_j \frac{\partial f}{\partial y_j} - \frac{\gamma(x_i - \mu_B)}{N(\sigma_B^2 + \epsilon)^{3/2}} \sum_j \frac{\partial f}{\partial y_j} (x_j - \mu_B)\]
The green expression above representing the direct path becomes the following code:
term1 = dout * gamma * inv_varThis is the direct pathway where \(x_i\) only affects \(y_i\) (not other outputs). The Kronecker delta \(\delta_{ij}\) from our derivation automatically becomes element-wise multiplication in the vectorized implementation:
dout[i] (\(i\)-th row)
contains \(\frac{\partial f}{\partial
y_i}\) (upstream gradient for output \(i\))gamma contains \(\gamma\) (scale parameter)inv_var contains \(\frac{1}{\sqrt{\sigma_B^2 + \epsilon}}\)
(inverse standard deviation)term1[i] with its corresponding dout[i]
term1[0] gets dout[0] * gamma * inv_var =
\(\frac{\partial f}{\partial y_0}
\frac{\gamma}{\sqrt{\sigma_B^2 + \epsilon}}\)term1[1] gets dout[1] * gamma * inv_var =
\(\frac{\partial f}{\partial y_1}
\frac{\gamma}{\sqrt{\sigma_B^2 + \epsilon}}\)term1[2] gets dout[2] * gamma * inv_var =
\(\frac{\partial f}{\partial y_2}
\frac{\gamma}{\sqrt{\sigma_B^2 + \epsilon}}\)\[\frac{df}{dx_i} = \frac{\partial f}{\partial y_i} \frac{\gamma}{\sqrt{\sigma_B^2 + \epsilon}} \textcolor{blue}{ - \frac{\gamma}{N\sqrt{\sigma_B^2 + \epsilon}} \sum_j \frac{\partial f}{\partial y_j}} - \frac{\gamma(x_i - \mu_B)}{N(\sigma_B^2 + \epsilon)^{3/2}} \sum_j \frac{\partial f}{\partial y_j} (x_j - \mu_B)\]
The blue expression above representing the mean path becomes the following code:
term2 = -gamma * (1. / N) * inv_var * np.sum(dout, axis=0)This captures how \(x_i\) affects all outputs through the shared mean \(\mu_B\):
np.sum(dout, axis=0) computes \(\sum_j \frac{\partial f}{\partial y_j}\)
(sum of upstream gradients across the batch). To understand this:
dout has shape (N, D) where each row
dout[j] contains \(\frac{\partial
f}{\partial y_j}\). When we sum along axis=0, we're
adding up all the rows:
dout[0] + dout[1] + ... + dout[N-1], which gives us exactly
\(\sum_j \frac{\partial f}{\partial
y_j}\) for each feature dimension.(D,), because np.sum
will collapse the columns of the (N,D) array, leaving one value per
feature dimension\[\frac{df}{dx_i} = \frac{\partial f}{\partial y_i} \frac{\gamma}{\sqrt{\sigma_B^2 + \epsilon}} - \frac{\gamma}{N\sqrt{\sigma_B^2 + \epsilon}} \sum_j \frac{\partial f}{\partial y_j} \textcolor{red}{- \frac{\gamma(x_i - \mu_B)}{N(\sigma_B^2 + \epsilon)^{3/2}} \sum_j \frac{\partial f}{\partial y_j} (x_j - \mu_B)}\]
Lastly, the red expression above representing the variance path becomes:
term3 = -gamma * (1. / N) * (x-mean) * inv_var**3 * np.sum(dout * (x - mean), axis=0)This captures how \(x_i\) affects all outputs through the shared variance \(\sigma_B^2\):
(x-mean) (outside the np.sum)
represents \((x_i - \mu_B)\) - the
distance of each specific input from the mean. This gives us a separate
value for each example in the batch.(x-mean) (inside the np.sum)
represents \((x_j - \mu_B)\) when
summed. Specifically, np.sum(dout * (x - mean), axis=0)
computes \(\sum_j \frac{\partial f}{\partial
y_j} (x_j - \mu_B)\) by summing across all examples:
dout[0]*(x[0]-mean) + dout[1]*(x[1]-mean) + ... + dout[N-1]*(x[N-1]-mean).inv_var**3 gives us \(\frac{1}{(\sigma_B^2 +
\epsilon)^{3/2}}\)\[\frac{df}{d\gamma} = \sum_i \frac{\partial f}{\partial y_i} \cdot \hat{x}_i\]
becomes
dgamma = np.sum(dout * x_hat, axis=0)This captures how each normalized input \(\hat{x}_i\) scales the effect of \(\gamma\) on the final loss:
dout * x_hat gives us \(\frac{\partial f}{\partial y_i} \cdot
\hat{x}_i\) for each example (element-wise)np.sum(..., axis=0) sums across the batch dimension,
giving us one gradient value per feature\[\frac{df}{d\beta} = \sum_i \frac{\partial f}{\partial y_i}\] becomes
dbeta = np.sum(dout, axis=0)dout contains \(\frac{\partial f}{\partial y_i}\) for each
outputnp.sum(..., axis=0) sums across the batch
dimensionIf you made it this far, thank you for reading, please reach out and let me know what you thought over email or X! While I was studying this material I found that keeping all the intermediate variables and their dependencies in my head was too hard and clouded my understanding of what I'm doing. Whereas I found that using the "three pathways" framing and applying the chain rule made it much easier to keep context of where I was in the derivation and what it meant, in english. I'm curious to hear what you think.
Good luck :)
Namra